Data Standards
Supporting standards throughout the data lifecycle
What are metadata?Metadata are contextual information about your experimental data, and are critical for the sharing and reuse of scientific data. |
What is FAIR for microbiome research?Making microbiome data Findable, Accessible, Interoperable, and Reusable (FAIR) promotes data stewardship. |
What is the data lifecycle for microbiome research?Data management best practices span from project start to end. Standards play a critical role in supporting the data lifecycle, and well-established standards like the Genomic Standards Consortium’s (GSC) Minimum Information about any (x) Sequence (MIxS) reporting standard and the Environment Ontology (EnvO) are key to microbiome data use and reuse. The NMDC aims to support the data lifecycle by providing tools and resources for how to follow best practices and use standards. The NMDC Metadata Documentation provides further details on how we are working across standards organizations and with the research community. |
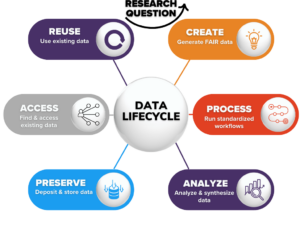 |