Standardized Bioinformatics Workflows
The NMDC provides accessible standardized workflows for processing microbiome omics data
Motivation
Microbiome datasets are often processed with different tools and pipelines, which presents challenges for reuse and cross-study comparisons. To address these challenges, the NMDC integrates production quality, open-source bioinformatics tools into accessible standardized workflows for processing omics data (e.g., metagenome, metatranscriptome, metaproteome, and metabolome data) to produce interoperable and reusable annotated data products. These workflows further the NMDC’s commitment to the FAIR data principles.
The NMDC workflows include bioinformatics tools developed by the Joint Genome Institute (JGI), Environmental Molecular Sciences Laboratory (EMSL), and Los Alamos National Laboratory (LANL), among others. The workflows developed and used in production at JGI and EMSL are used to process thousands of datasets annually and have been extensively benchmarked to ensure the generation of high-quality data.
The NMDC offers a user-friendly web interface, NMDC EDGE, where users can input their microbiome data, run the standardized NMDC workflows, and obtain summaries, output files, and visualizations of their results. NMDC EDGE is run using NMDC computing resources, and is free and open for researchers to process their omics data.
The NMDC workflows are also publicly available through GitHub and DockerHub as standalone, containerized workflows, offering a unique opportunity for any institute or individual to obtain, install, and run the workflows in their own environments.
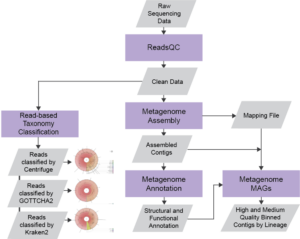
The NMDC Metagenome Workflow
Documentation
The NMDC documentation provides additional information on each workflow, their standardized parameters, any associated databases, versions, and the tools associated with each workflow.
Tutorials
Tutorial videos and user guides are available in the ‘Tutorials’ section of NMDC EDGE. User guides are available in English, Spanish, and French. These walk users through how to run each of the NMDC workflows in NMDC EDGE, and explain required input file formats and the outputs that are generated.
Feedback
For troubleshooting and questions about the workflows, users can email the team directly at nmdc-edge@lanl.gov with their issue and project name. To provide comments about the workflows that do not need follow-up from the team, a Google form is linked on the home page of the NMDC EDGE website. Our team works to incorporate as much user feedback as possible and we highly value user input and suggestions. Beta testing opportunities are also announced through the NMDC newsletter, “The Microbiome Standard”, on the NMDC User Research webpage, and communications channels such as the NMDC Community Slack. To participate, sign up here and the team will reach out when a beta testing round opens.