Connecting data, people, and ideas
Our Story
Building a FAIR microbiome data sharing network, through infrastructure, data standards, and community building, to address pressing challenges in environmental science.
In 2016, the White House Office of Science and Technology Policy (OSTP), in collaboration with federal agencies and private-sector stakeholders, launched the National Microbiome Initiative focused on three main priorities: supporting interdisciplinary research, developing platform technologies, and expanding the microbiome workforce. This spurred a call to action for microbiome data science, led by Nikos Kyrpides, Natalia Ivanova, and Emiley Eloe-Fadrosh. A small workshop convened in early 2017 at the Department of Energy’s Joint Genome Institute focused on developing a vision for microbiome data science to address gaps in existing infrastructure.
Working from a collaborative vision for a microbiome data infrastructure that was outlined at the workshop, an open-invitation town hall was organized at the 2017 ASM Microbe conference to gauge support and solicit input from the microbiome research community. Stakeholders from all facets of microbiome science filled the standing-room-only meeting and signaled a collective eagerness for concerted data infrastructure solutions. In November 2017, a stakeholder workshop hosted by the American Society for Microbiology brought together representatives from academia, industry, government, and philanthropic funding agencies to conceptualize the National Microbiome Data Collaborative (NMDC). These efforts serve as the foundation of a community-driven national effort aimed to develop standards, processes, and infrastructure for an integrated microbiome data ecosystem.
Following these program development activities, the FY19 Energy and Water Appropriations Bill included $10 million to “begin establishment of a national microbiome database.” In January 2019, Berkeley Lab was formally tasked by the DOE Office of Biological and Environmental Research to develop a pilot NMDC as a non-competed program. The NMDC was initiated in July 2019 as a pilot project led by Berkeley Lab, in partnership with Los Alamos National Laboratory (LANL), Pacific Northwest National Laboratory (PNNL) and Oak Ridge National Laboratory (ORNL).
The NMDC pilot focused on developing core capabilities and resources centered on standards and metadata; bioinformatic workflows; data integration and access; and community engagement. During the pilot phase, we also developed strategic partnerships with DOE’s Environmental Systems Science Data Infrastructure for a Virtual Ecosystem (ESS-DIVE), DOE’s Systems Biology Knowledgebase (KBase), and NSF’s National Ecological Observatory Network (NEON). To advance the NMDC beyond the pilot phase, we were invited to prepare a 3-year plan in 2022 that leveraged lessons learned and an ambitious framework for collaborative, interdisciplinary data infrastructure to support integrative science.
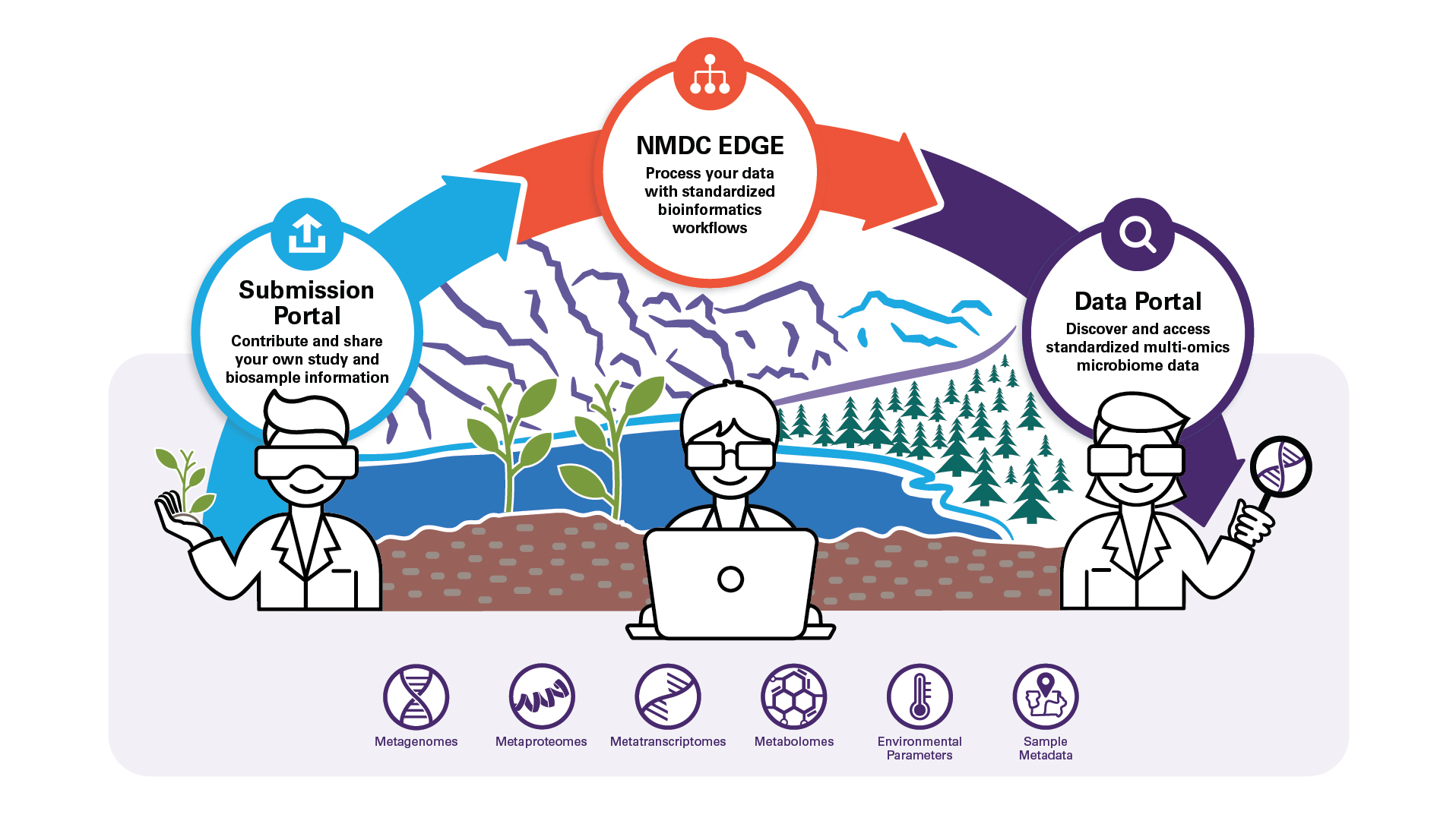
We use proven approaches and new innovations in distributed data infrastructure and linked data technologies to support new ways for researchers to ask questions about how genes, individual microbes, and communities are associated with environmental processes. Engagement and user research throughout the development lifecycle ensures we are providing resources which are essential to advancing microbiome research. The NMDC aims to tackle existing gaps in environmental microbiome research to broadly support data, information, and knowledge access through the NMDC products driven by community needs.
Enabling inclusive and interdisciplinary environmental microbiome science by connecting data, people, and ideas.
Funding:

How to cite the NMDC:
Data Portal
Ambassador Program
Program Launch