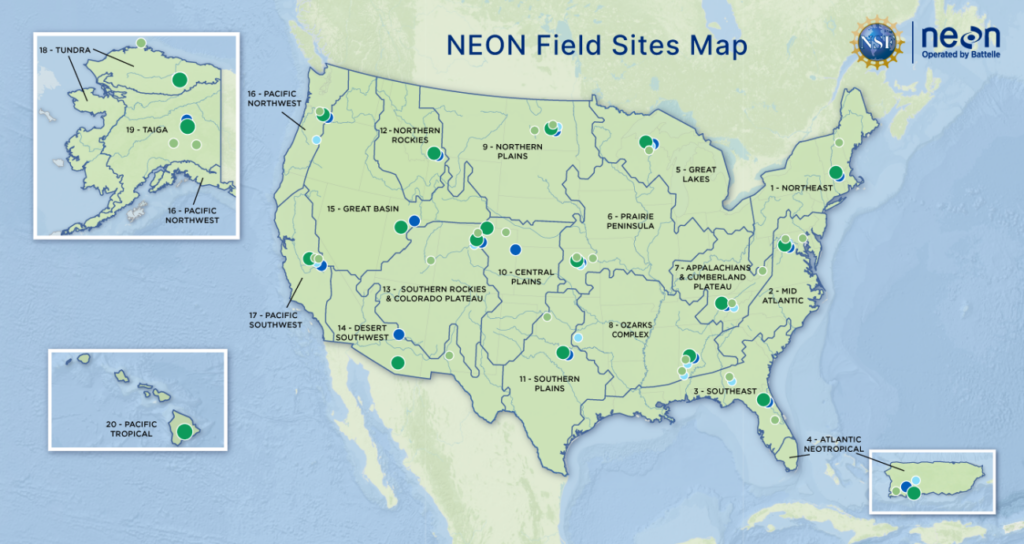
NEON field sites and ecoclimatic Domains. NEON (National Ecological Observatory Network). https://www.neonscience.org/field-sites/about-field-sites
Even the minute movements of tiny organisms within an invisible microbiome impact how plants and animals interact on land and in the water. Understanding the interplay between these micro and macro worlds allows researchers to comprehend how environmental changes echo throughout every part of our world. The National Microbiome Data Collaborative (NMDC) actively collaborates with research teams and consortiums to collect and share standardized microbiome data to enable further discovery and access. The National Ecological Observatory Network (NEON) is an example of one of these collaborations. The NEON is a continental-scale observation facility operated by Battelle and fully funded by National Science Foundation (NSF). The NSF created NEON to collect long-term open-access ecological data to understand how U.S. ecosystems are changing. NEON provides more than 180 data products, including metagenomic sequencing results, from diverse field sites around North America, freely available on their website, neonscience.org.
There are clear links between the environmental microbiome data supported through the NMDC and the ecological data from NEON. Since a memorandum of understanding was established between the DOE and NSF in 2021, NEON has aligned with the NMDC to maximize collaborative efforts to leverage scientific investments for microbiome data standards, sharing data processing approaches, and coordination of data access and availability through NEON and NMDC resources. NEON and the NMDC have frequently collaborated to provide users access to NEON data and start conversations about metadata standardization. The NMDC ingests multi-omics data and contextual information from NEON, adding them to the ever-expanding information available in the NMDC Data Portal. Currently, data from over 4,000 NEON samples exists within the Data Portal.
Additionally, NEON staff have served as panelists and co-hosts for several NMDC events. At the June American Society for Microbiology Microbe conference, Battelle bioinformatician Dr. Hugh Cross discussed the challenges of studying the interplay between climate change and its impacts on environmental systems. Cross and NEON Science Lead Dr. Kate Thibault, also co-led a workshop about FAIR data with NMDC team members at the Ecological Society of America conference in August. They also collaborated with NMDC team members on a special session on accessing environmental and ecological omics data. Researchers can use NEON and NMDC’s resources to find additional data related to their research and analyze standardized datasets.
Thanks to new support through the Joint Genome Institute’s (JGI) Community Science Program (CSP), the NEON team is poised to bring even more metagenomics data to support continental-scale analyses of samples from terrestrial and aquatic microbial communities. A new CSP leverages the NEON and NMDC partnership to advance researchers’ understanding of terrestrial and aquatic microbial communities. The Department of Energy’s Biological and Environmental Research Program (BER), which supports both the NMDC and JGI, seeks to help researchers cultivate an understanding of biological, biogeochemical, and physical processes that span from molecular and genomics-controlled scales to the regional and global scales that govern changes in watershed dynamics, climate, and the earth system. Similarly, NEON aims to provide high-quality, national-scale critical data and samples that can be widely accessible by all. In their proposal, NEON PIs Cross and Thibault outlined a plan to make more data available to researchers by utilizing JGI’s unique resources and technology. NMDC team members Julia Kelliher (Los Alamos National Laboratory) and Montana Smith (Pacific Northwest National Laboratory) are co-PIs on the project to support metadata harmonization and data integration.
Leveraging the latest short-read sequencing technology with Illumina’s NovaSeq X, the JGI will sequence over 1,000 NEON samples spanning 81 field sites, including 47 terrestrial and 34 aquatic sites, throughout 20 statistically derived, ecoclimatic domains of the US, including Alaska, Hawaii, and Puerto Rico, with sites varying from deserts to rainforests. Data will be generated and processed using JGI standardized analysis pipelines (assembly, annotation, and metagenome-assembled genome [MAG] binning). Then, the NMDC team will directly pull samples and data into the NMDC’s Data Portal and augment with reads-based analyses. This CSP will significantly increase the sequencing outputs of NEON samples, along with creating even more broadly sharable microbiome data that enables new scientific research and enhanced institutional coordination. Thanks to this collaboration, each sample will be processed with significantly deeper sequencing, generating a more complete picture of the microbes within these samples. The NMDC team is excited to continue working with the NEON team to help ecologists, microbiologists, and other researchers learn more about the dynamics of microbes in freshwater and soils throughout the US across space and time. Understanding how these dynamics are influenced by changes in land use and climate is critical to our shared goal of maintaining the health and productivity of ecosystems.
Join our vision
Want more info? Or to be an NMDC Champion? Subscribe to be the first to know about the latest news and developments.
